This is the first post in a two-part series. Read Part 2 here.
Around the same time that Alan Turing was shaping his theories of machine intelligence in Manchester, another future giant of the computing world, Douglas Engelbart, was developing an alternative computing paradigm over 5,000 miles away in the Bay Area.
Engelbart believed that computers, with their ability to synthesize and manipulate vast quantities of information, should help humans solve problems, rather than remove them from the problem-solving loop. This ideology is now known as augmented intelligence. Engelbart’s contributions to the field (both as a PhD student at UC Berkeley and at SRI in the decades after) were perhaps best exemplified through “The Mother of All Demos” in 1968, where he unveiled for the first time many of the computing features we now take for granted — the mouse, GUIs, hyperlinks, word processing, version control, and even video conferencing — in a single demonstration.
Although it’s enticing to think about artificial intelligence passing human equivalency tests like Turing’s Imitation Game (or maybe something more sophisticated for today’s generalist AI models), we really should be thinking about how Engelbart’s ideas translate to our modern AI era. Put another way, how do we build the next Mother of All Demos?
In reality, the next mother of all demos will be much more than just a demonstration. We already have the ingredients — a whole new set of AI tools — so now we need to think about how these ingredients can help us reimagine and redesign our existing workflows and user experiences. In doing so, we can usher in a new class of AI-native experiences for search, scientific research, game design, and more.
The Mother of All Demos in the age of deep learning
If we’re building a version of the Mother of All Demos in the deep learning era, we could begin with these models and tools. Although research labs release new models seemingly every week, these are a great starting point to begin rethinking how we interact with technology:
GPT-3
The latest in OpenAI’s GPT series, GPT-3 is a 175-billion parameter language model that is trained on practically all of the text that exists on the Internet. Once trained, GPT-3 can generate coherent text for any topic (even in the style of particular writers or authors), summarize passages of text, and translate text into different languages. OpenAI also recently released a follow-up, InstructGPT, that incorporates human feedback to reduce harmful or biased outputs.
Copilot
Github’s Copilot takes “translating text into different languages” to the programming world. Built on top of OpenAI’s Codex — think GPT-3, but trained on words and code — Copilot is an “AI pair programmer” that will generate anything from individual lines to whole functions of code, based on a docstring describing what the code is supposed to do. Recently, DeepMind released a competing code-writing model, AlphaCode, that solves coding challenges from popular programming competitions.
DALL-E
While GPT-3 and Copilot generate text from text, DALL-E learns to generate images from text. In fact, it’s the same model architecture as the GPT, just trained on text-image pairs instead of text alone. Though DALL-E’s 12-billion parameters are an order of magnitude fewer than GPT-3, the final product is still quite powerful — DALL-E can generate anything from playful sketches to photorealistic images, and even create drawings that include concepts that aren’t found in DALL-E’s training data (or anywhere, for that matter). OpenAI recently released GLIDE as a lighter-weight (but equally performant) version of DALL-E.
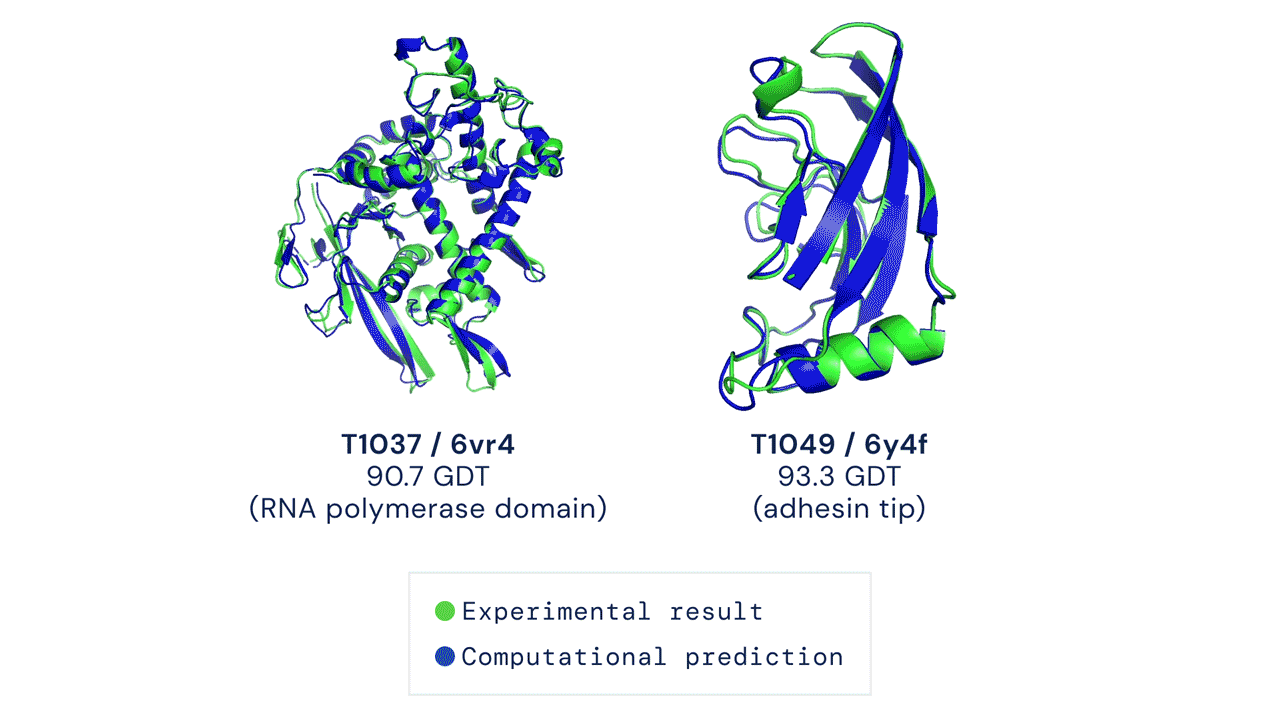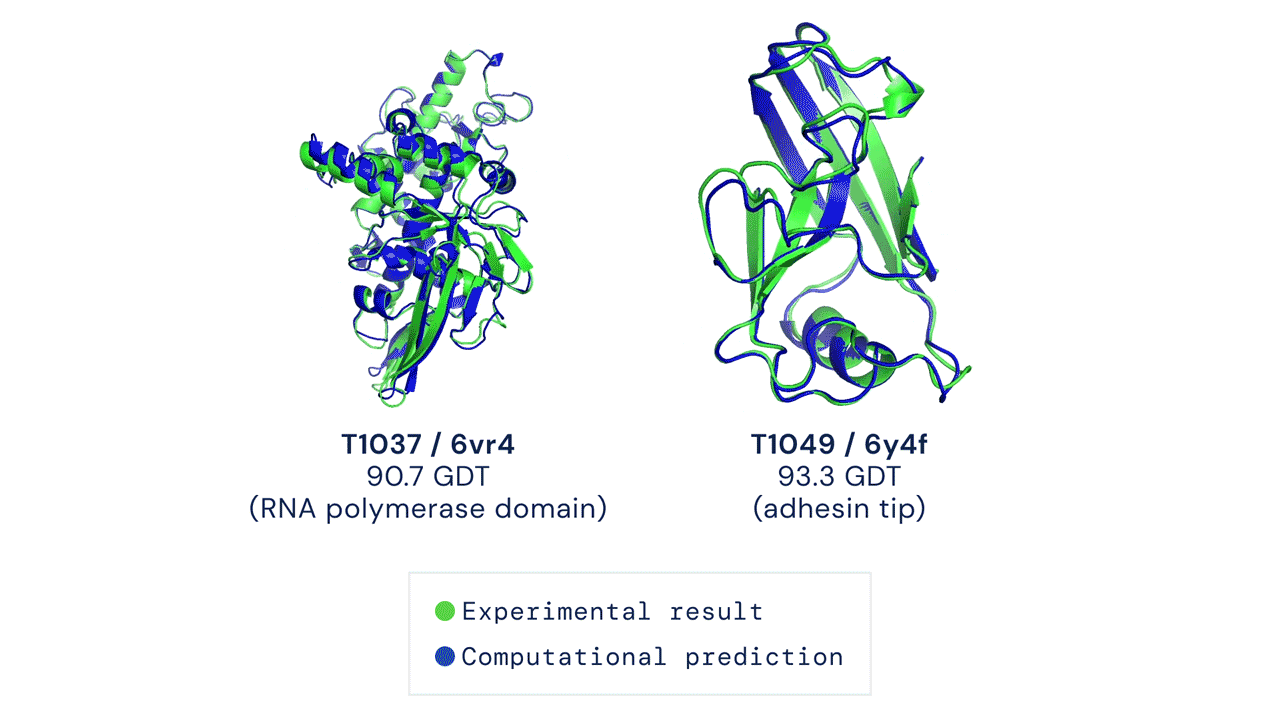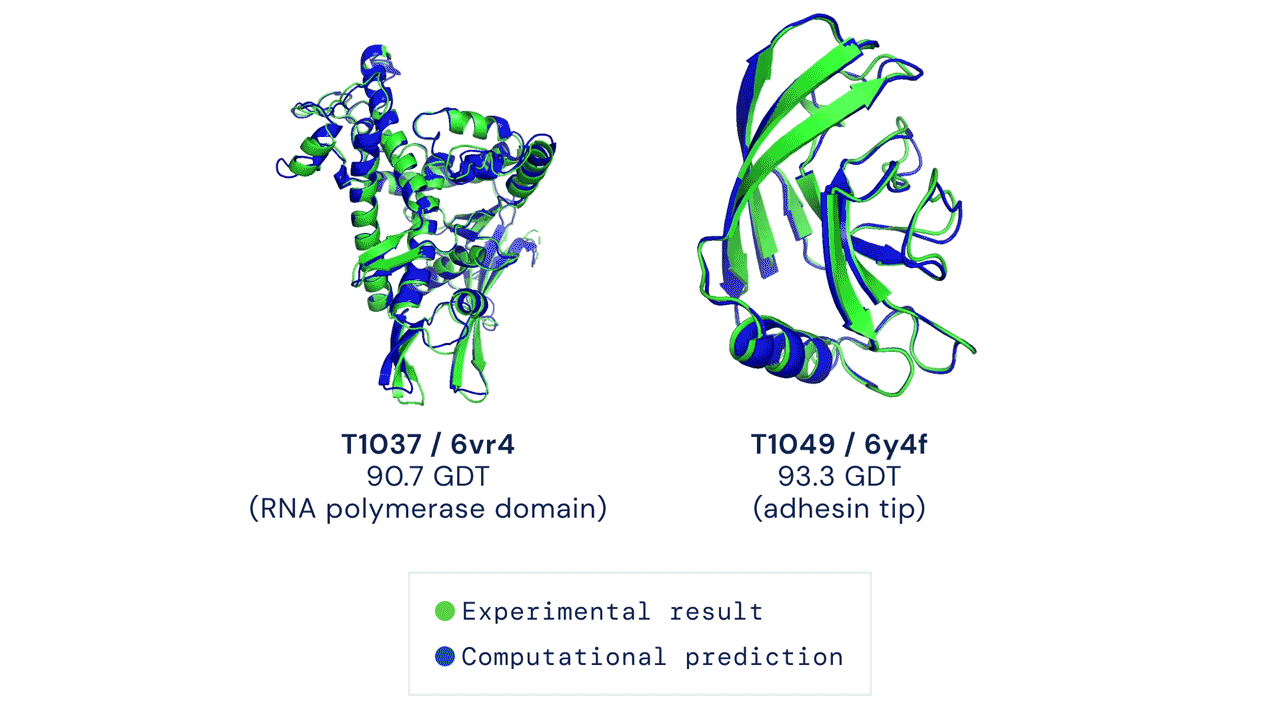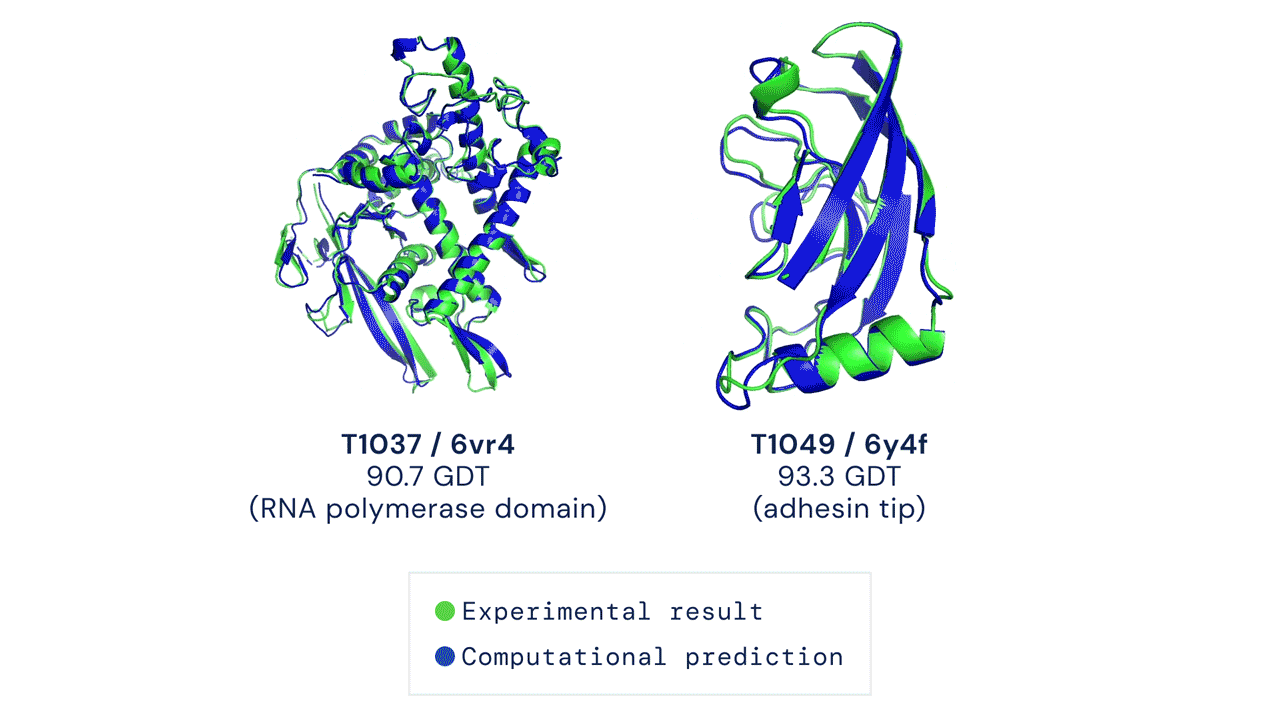
AlphaFold
DeepMind’s AlphaFold differs from the prior models in that it is not intended to be general purpose, but instead solves one very specific problem: protein-folding. AlphaFold has learned to predict the 3D structure of proteins from their component amino acid parts and, using around 170,000 known protein structures as training data, can now predict up to 98.5% of the proteins that exist in the human genome. Thanks to the ubiquity of proteins in biology and the relationship between protein structure and protein function, predicting protein structures accurately gives AlphaFold enormous potential across a wide range of industries and applications.
With that introduction to these popular AI models, here are some thoughts on how we could use them to improve and, really, reimagine how we perform certain tasks, processes, and even jobs.
A curated and informed search experience
“There is a growing mountain of research. But there is increased evidence that we are being bogged down today as specialization extends. The investigator is staggered by the findings and conclusions of thousands of other workers.”
— As We May Think, Vannevar Bush, 1945
At the time of writing the above, Vannevar Bush, the academic turned wartime scientific adviser to Presidents Franklin Roosevelt and Harry Truman (and a substantial influence on Engelbart), was concerned that the “information explosion” provoked by the frenzy of R&D during WWII would impact progress in the post-war scientific community. Only by developing new tools to combat information overload, Bush suggested, could scientists be freed to pursue “objectives worthy of their best.”
The web takes things to a whole new level. While scientists in Bush’s era communicated their research through a handful of physically bound journals in each research area, the web has led to an exponential explosion of scientific research that is published primarily online (open-access sites like Arxiv are a good example of this). A recent study suggests that the academic community accounts for over 8 million new articles per year — a far cry from the tens of thousands of articles published in the 1940s.
And remember, this is just an estimate of the number of academic articles. As of 2016, the largest newspapers in the United States were publishing hundreds of pieces per day each. Combine academic articles and newspaper articles, and you haven’t even scratched the surface of what’s available on the web! If we’re going to continue producing so much, we’re going to need a better search experience.
As an example of the challenges posed by today’s information overload, let’s work through a little thought experiment. Say that I wanted help writing this article and turned to Google by searching “article on augmented intelligence and deep learning”:
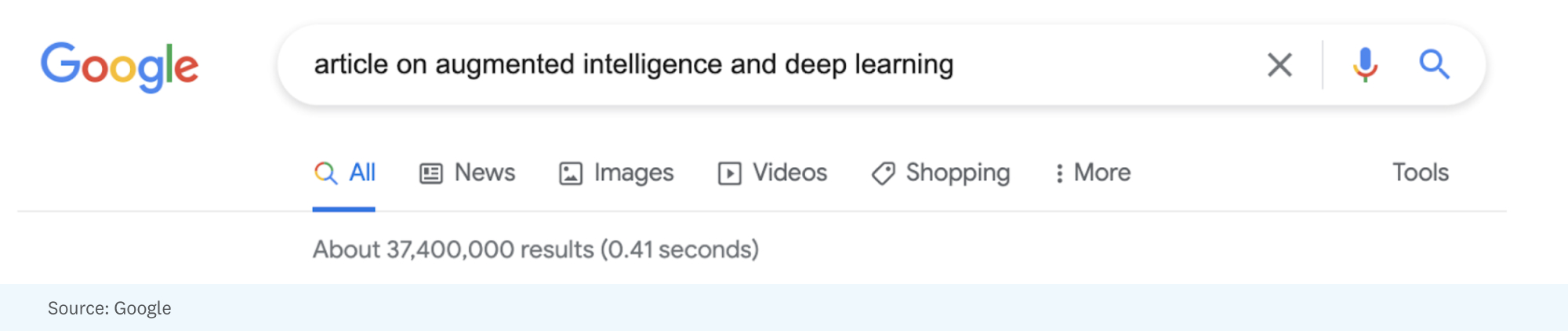
Those 37.4 million results might be a mix of articles, op-eds, blog posts, social media and message board posts, videos, and research publications. The challenge is picking the most relevant (5? 10? 100?) results, summarizing them, and then synthesizing them into suggestions, possible references, and maybe even candidate sentences/paragraphs for an article that readers of Future might find interesting.
Dealing with this firehose of information is a problem of dimensionality reduction — distilling hundreds of thousands (or millions) of versions of something into a small, workable set. Google historically did this by showing you the most relevant pages first, where “relevance” is determined by PageRank — Larry Page’s famous algorithm that combines semantic similarity between a page and a user’s query with the number of links pointing to a page.
However, proper dimensionality reduction requires a search that goes beyond counting references. For example, you can imagine a number of different themes that could underpin an article on this topic, such as AGI vs. augmented intelligence through a purely economic lens, or a technical deep dive on how AI and augmented intelligence systems are built. You can also imagine a spectrum of sentiment ranging from strongly pro-AGI or anti-Engelbart, to pieces denouncing AGI and stamping dates on the next AI winter. This article fits somewhere in the middle.

To make the most of the growing mountains of content available to us on the web, we need to be able to associate examples across all of these axes separately. Moreover, we need to find the right number of these buckets to show the user. Themes, opinions, and sentiment are a good start, but are there other ways of categorizing content that we are missing? Too few buckets may limit a user’s options; too many buckets just kicks the information-overload can down the road.
Fortunately, GPT-3 can help us out. GPT-3 is a general-purpose language model, so it can argue any point, for any topic, in any number of ways. It can also summarize any passage of text and extract key statistics. It’s entirely possible that we can use GPT-3 to do a first pass over content, ranking each piece over multiple dimensions like those mentioned above, and categorizing accordingly (presumably through a redesigned search interface). If we’re doing research for an article, as in my prior example, we may also be able to lean on GPT-3 to highlight main points and other information to include, based on the author’s goals.
A GitHub for protein development
AlphaFold is also an interesting example here — but as a source of valuable information that could really benefit from a new search experience. The AlphaFold Protein Structure Database contains around 360,000 predicted protein structures and DeepMind plans to add around 100 million (!!) more protein structures this calendar year. Additionally, many experts have touted the potential of this database for de novo protein design — the process of algorithmically creating new proteins that do not appear naturally but adhere to natural folding laws. The applications of de novo proteins are profound — new proteins will help us develop better vaccines, fight cancer, and break down materials that harm the environment — but once we open the floodgates, brute-force strategies for discovery and experimentation just won’t cut it.
To make this de novo landscape actionable, and to operationalize AlphaFold in general, we’ll need to be able to search for and prioritize proteins based on their structure and possible function. We’ll also need to update and redesign other fundamental computational tools like version control and backtesting.
For example, imagine you are targeting a class of similarly structured de novo proteins that might be effective for breaking down carbon in the atmosphere. AlphaFold gives you 3D structure, but to take this (possibly large) group of protein candidates from simulation to reality you’ll have to synthesize each individual protein and collect experimental results to test its feasibility. Lab experiments are slow and expensive, though, so starting from scratch for each new experiment isn’t an option.
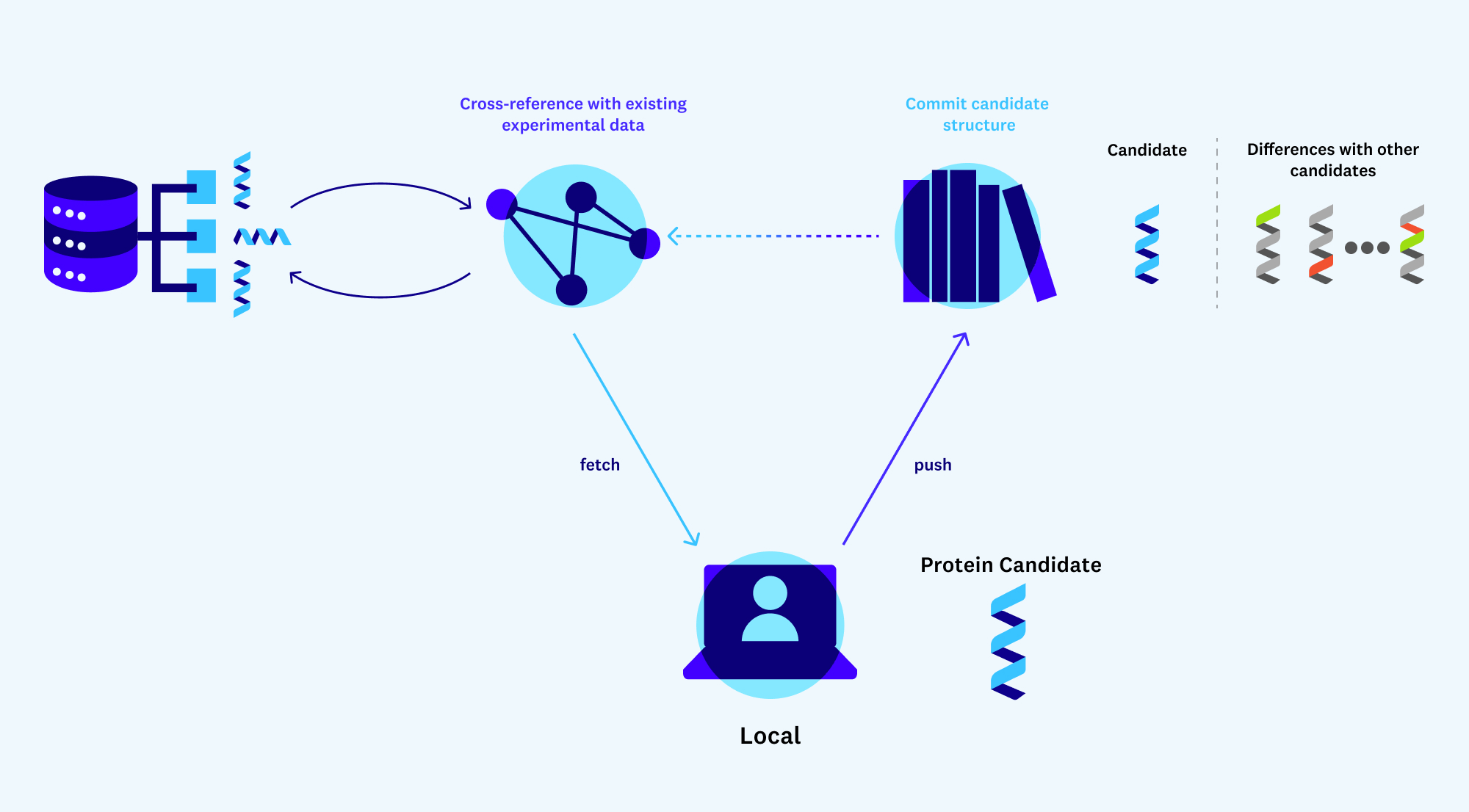
In some cases, there might be experimental results for a similar, naturally existing protein that has been conducted manually in a lab somewhere. You’ll need a way to streamline how results for your new proteins might compare to these existing results. Even better would be a GitHub-like service for protein design that allows experimentalists to store the diffs of protein candidates and any experimental results that exist for them. This would unlock the benefits of open source and reduce the barriers to entry of working with novel proteins.
Changing tone like we change fonts
Before windows and GUIs, there was the terminal. Before the terminal, there were punch cards. Generative AI models like GPT-3, AlphaFold, and DALL-E are currently having their “terminal” moment. Interacting with them does not require manually punching holes in cards, but it does require working knowledge of programming, APIs, and at least a basic grasp of how the model works under the hood. There is a massive opportunity to use these models to reimagine how we interact with and visualize workflows for the problems we solve.
Consider GPT-3, which generates coherent text but lacks in common sense and its ability to invent entirely new ideas from existing data sources. (The latter problem was referred to by Turing as “Lady Lovelace’s objection,” in reference to Ada Lovelace’s famous assertion that computers have “no pretensions whatever to originate anything.”) We should probably not, then, redesign word processors to look and function like the Google homepage — all you need is a little text field for input and out comes your essay, letter, or poem! This technically makes the writing process easier, but does not make writing (or communication) any better.
Instead, we should seize the opportunity to redesign the writing process around the human strengths of ideation and revision. We can replace blank pages and blinking cursors with a canvas for creating and rearranging webs of concepts, themes, and thought bubbles, and then let GPT take it from there. We can build tools for increasing or decreasing the sentiment of a sentence or modifying the prosaic style of a paragraph with the same ease with which we change fonts today.
A recent project out of Stanford has produced an exciting GPT-3-backed tool called CoAuthor that represents a first step toward some of the augmentation possibilities we’ve discussed. CoAuthor allows writers to request suggestions from GPT-3, displaying the small snippets of text in a similar way to how autocomplete suggestions are shown on smartphone keyboards. Here is an example of an interactive writing session with CoAuthor:

Interactive experimentation for complex science
You could envision similar modifications for AlphaFold. Although AlphaFold is capable of generating 3D protein structures with unprecedented accuracy, it solves only half of the large, hairy problem that is understanding protein behavior. By understanding a protein’s structure, we have a clue to its function. But protein behavior is a function of both structure and context. Proteins fold into their structures under specific conditions, such as when they bind with other proteins/substrates, react to chemicals, or interact with surrounding molecules.
AlphaFold currently ignores these conditions. Moreover, understanding information that is immediately actionable like, say, how a new drug is likely to interact with a particular protein (and at what binding sites) requires finer-grain mapping than AlphaFold can currently provide. (Future iterations of AlphaFold will target protein-protein complexes, which will help somewhat with this problem.)
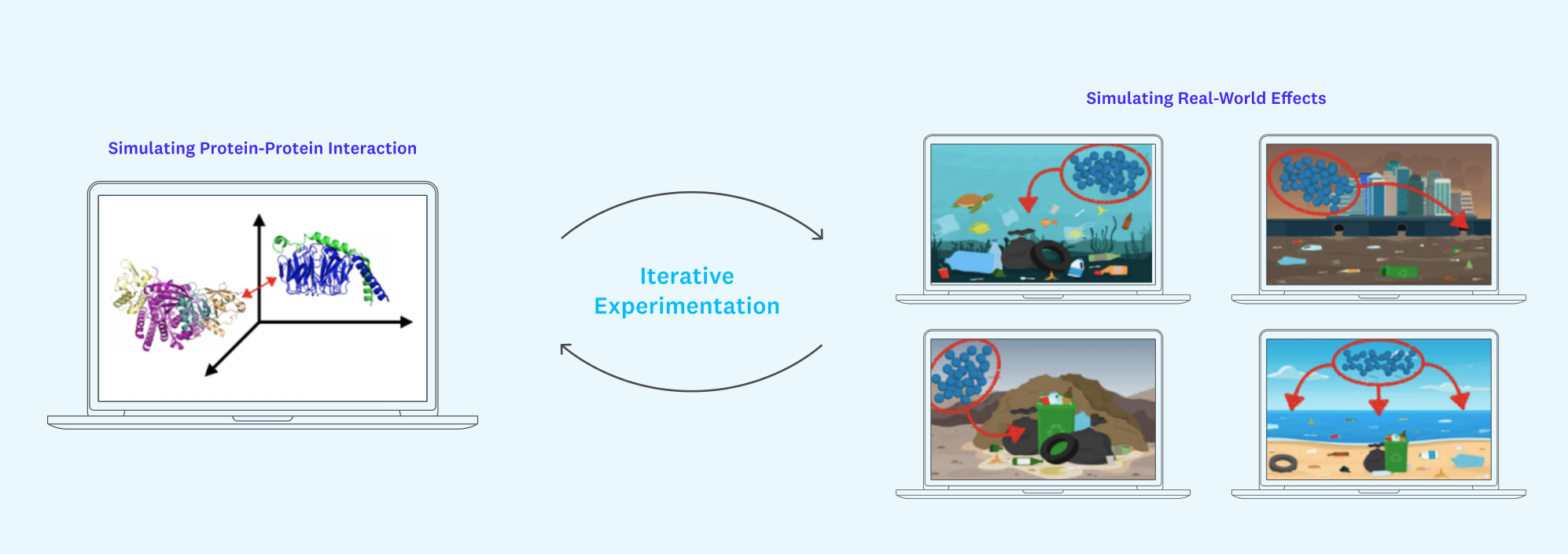
It will be crucial to be able to fill in this context in a streamlined way that is accessible to both protein experts and those without PhDs in biology. Unity is a great case study here. By providing fundamental infrastructure for rendering 3D graphics and simulating physical phenomena like gravity (and iterating on these ideas), Unity has played a pivotal role in accelerating progress for industries like gaming, self-driving vehicles, filmmaking, architecture/construction, and more.
Creating plug-and-play, Unity-style scene building with an encoding of what we know about protein-protein and protein-drug interactions under the hood might just allow us to generate large-scale simulations of how our target proteins might interact with, say, a mountain of plastic bottles or carbon-filled air currents over the Pacific Ocean. Such a tool would speed up basic tests of validity for protein use cases, filling in missing context and, crucially, lower the bar for less technical users to iterate on new ideas.
In this part, we’ve taken a first look at how we can reimagine existing workflows through the lens of recent advances in AI and deep learning. But there is a common thread in each of these use cases — they all take the model as is. In Part 2, we’ll explore a new set of tools and processes for interacting with these models, that make the models themselves more effective.
